Getting data into R
Note: This first appeared in Biometric Bulletin Volume 33, Issue 3, Software Corner.
One of the biggest hurdles that students and novice R users have is importing data into R. A number of recent packages have made this process easier and, importantly in the age of big data, faster. Most of the packages outlined below are part of Hadley Wickham’s tidyverse and owe their speed to calling C or C++ libraries from R.
The readr package
The readr package provides new functions for importing tabular data into R. Specifically, the functions read_table(), read_csv(), read_delim() are intended as fast (around 10 times faster) replacements for the base R read.table(), read.csv(), read.delim(). The readr functions do not convert strings to factors by default, are able to parse dates and times and can automatically determine the data types in each column (it does this by parsing the first 1000 rows). You can even import compressed files and they will be automatically decompressed and read into R. There is also file writing functionality, with write_csv(), write_tsv() and write_delim(). If you want even more speediness in your data importing, you might also consider the fread() function from the data.table package.
The readxl package
It used to be that the most reliable way to get data from Excel into R was to save it as a tab (or comma) delimited text file. While there are alternatives, such as the xlsx, XLConnect and gdata packages, that have other features (e.g. the ability to write excel files), if all you need is an easy way to import tabular data from xls and xlsx formats files, then the readxl package is for you. Importantly it has no external dependencies, so is very straightforward to install and use on all platforms. The syntax is very similar to the readr package functions, for example you specify the file name and the sheet of interest, read_excel("spreadsheet.xlsx", sheet = "data")
The haven package
The haven package provides functions for importing from SAS, SPSS and Stata file formats, read_sas(), read_sav()and read_dta(). This functionality is similar to that available in the base R foreign package but is often faster, can read SAS7BDAT files and formats, works with Stata 14 and 14 files. Following is the code to read SAS7BDAT.
install.packages("haven")
library(haven)
dat = read_sas("path to file", "path to formats catalog")The returned object will be a data frame where SAS variable labels are attached as an attribute to each variable. When a variable is attached to a format in SAS and the formats are stored in a library, its path also needs to be supplied. Missing values in numeric variables should be seamlessly converted. Missing values in character variables are converted to the empty string. To convert empty strings to missing values, use zap_empty(), for example,
dat$x1 = zap_empty(dat$x1)SAS, Stata and SPSS all have the notion of a “labelled”” variable. These are similar to categorical factor variables in R, but integer, numeric and character vectors can be labelled and not every value must be associated with a label. To turn a labelled variable into a standard factor R variable use the as_factor() function,
dat$facvar = as_factor(dat$facvar)The haven package is under active development and becoming increasingly robust. If you have difficulties loading a file, try using the development version on GitHub:
devtools::install_github("hadley/haven")For example, consider the National Youth Tobacco Survey (NYTS) from the CDC website. After downloading the files (and installing the development version from GitHub) the data can be imported into R using
devtools::install_github("hadley/haven")
require(haven)
x = read_sas("nyts2014_dataset.sas7bdat","nyts2014_formats.sas7bcat")
# convert qn1 to a factor:
x$qn1 = as_factor(x$qn1)
View(x)The rio package
The rio package describes itself as “a Swiss army knife for data I/O”. It unifies many of the above methods by providing the wrapper function import() that takes as an input the path to a data file. It then uses the file extension to determine the file type and imports the data into R. The one function can be used to import standard text files, RData, JSON, Stata, SPSS, Excel, SAS, XML, Minitab and many more. There is an analogous export() function that allows users to similarly easily export data to various file types.
Using RStudio to import data
Recent versions of RStudio allow users to import various file types via a graphical user interface, which is perfect for novice users and experts alike as they get used to the new functions and customization options. Once you’ve clicked through the various options, it will output the required code at the console so that you can see exactly what was done to get the data in and edit the code as necessary for next time.
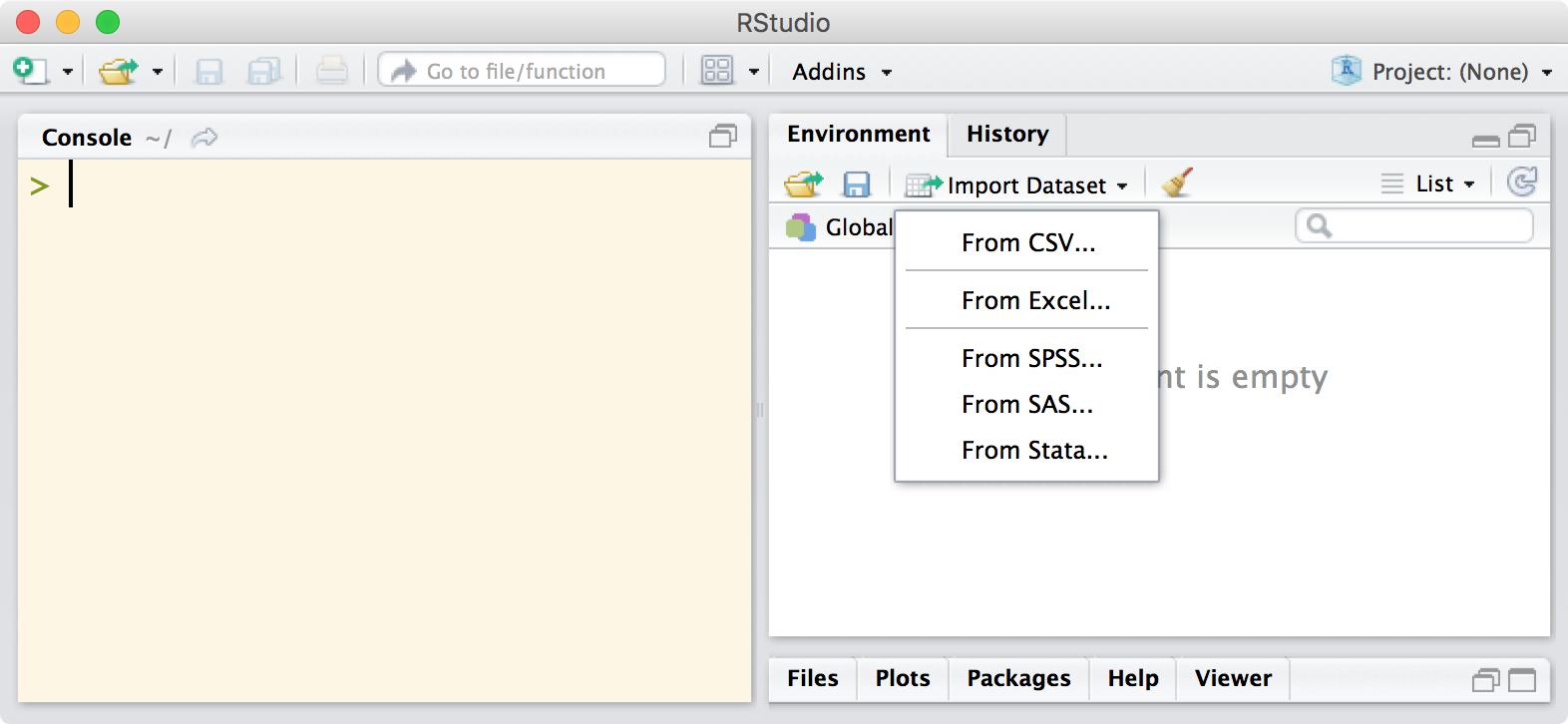
The team at RStudio have put together a webinar on getting data into R which is well worth watching.
Once you’ve got your data into R, you’ll probably need to restructure it in some way prior to analysis. To help with this, you may want to take a look at the tidyr package provides a suite of functions to get your data set in a standardized format, such that each observation is a row, each variable is a column and there are no data in the labels.